ANOVA also known as Analysis of Variance is a powerful statistical method to test a hypothesis involving more than two groups (also known as treatments). However, ANOVA is limited in providing a detailed insights between different treatments or groups, and this is where, Tukey (T) test also known as T-test comes in to play. In this tutorial, I will show how to prepare input files and run ANOVA and Tukey test in R software. For detailed information on ANOVA and R, please read this article at this
link.
Step 1.0 Download and install R software and R studio
- Download and install the latest version of the R software from this link
- Download and install R studio from this link
- Finally, install the library qtl in R
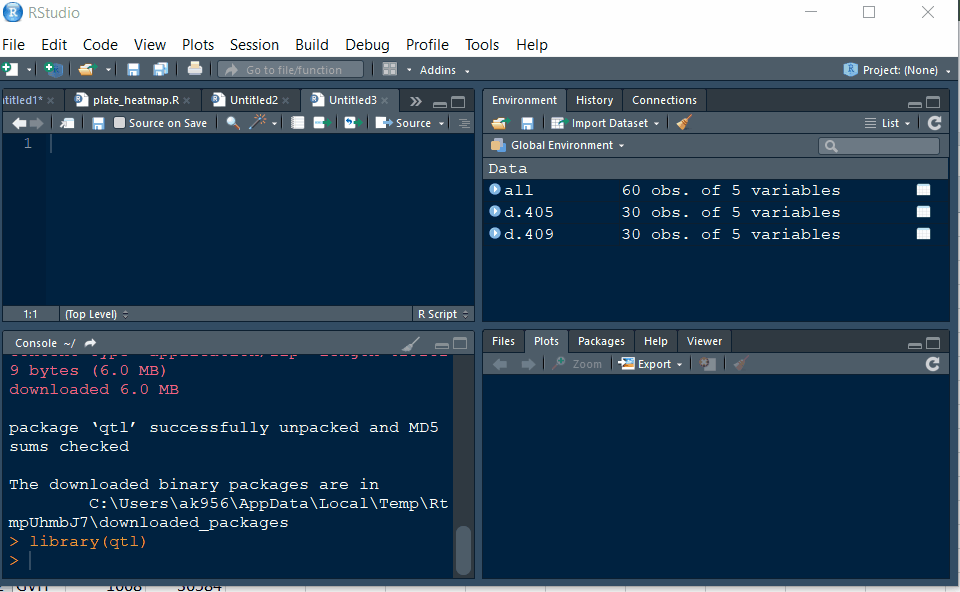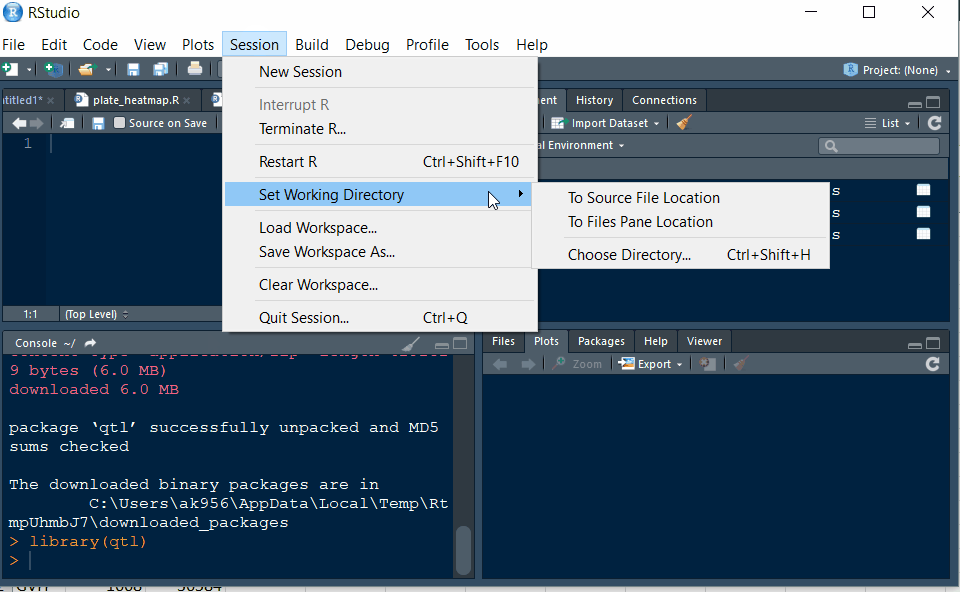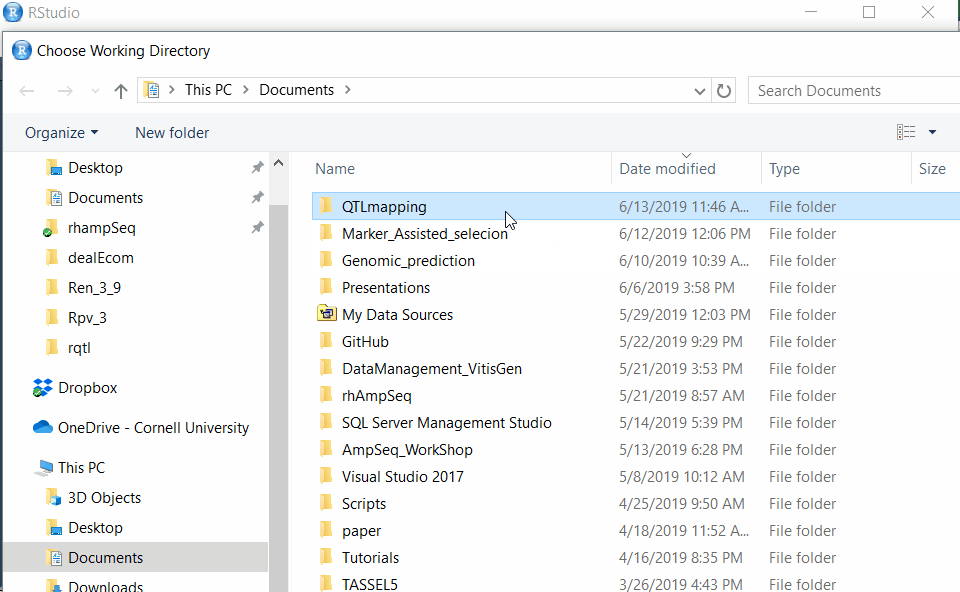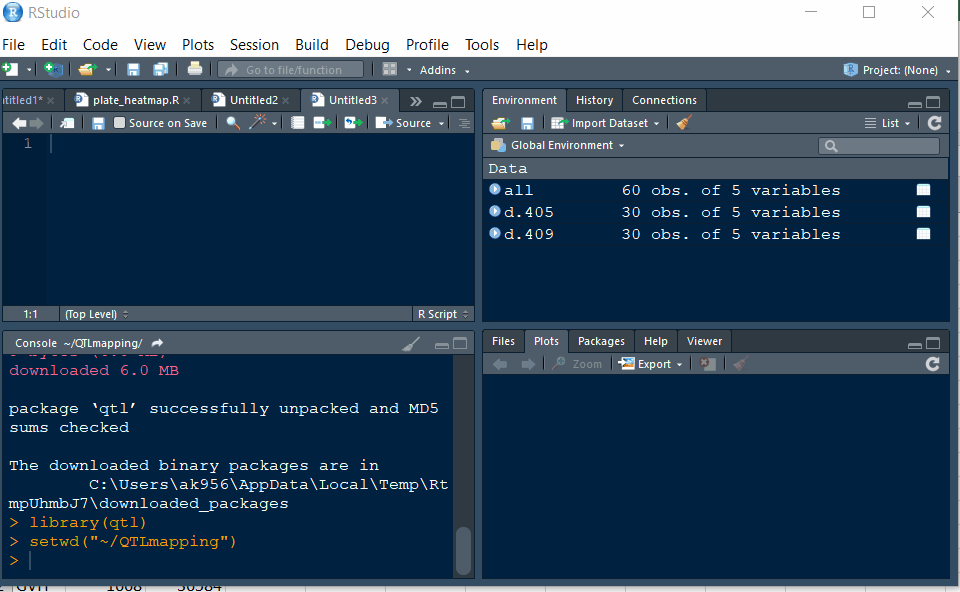
Step 1.2 - Setup working directory following the below steps:
Step 1.3: Preparing the Input file
Create an input file as shown in the example below:

Step 2: Run ANOVA in R
2.1 Import R package
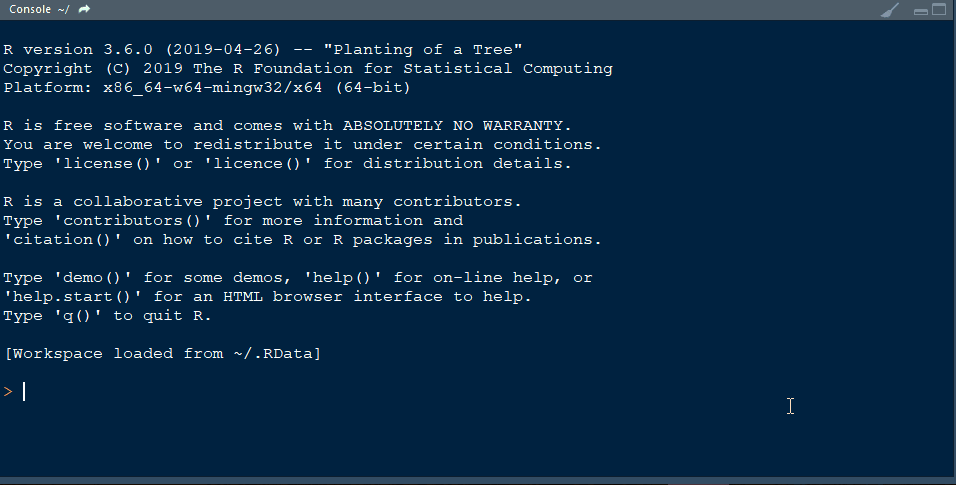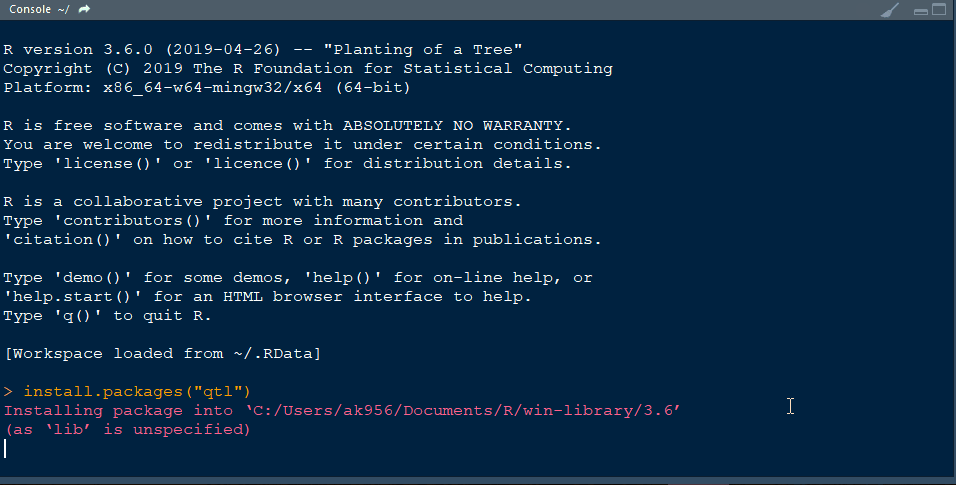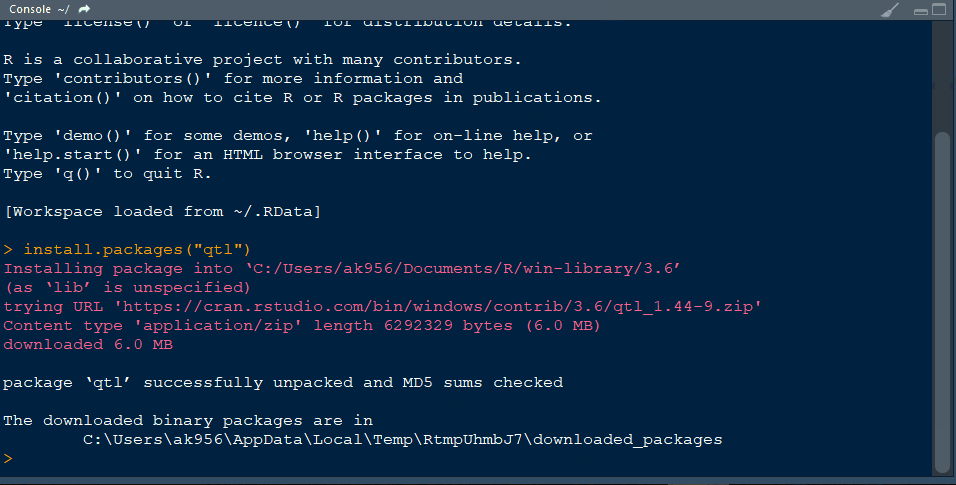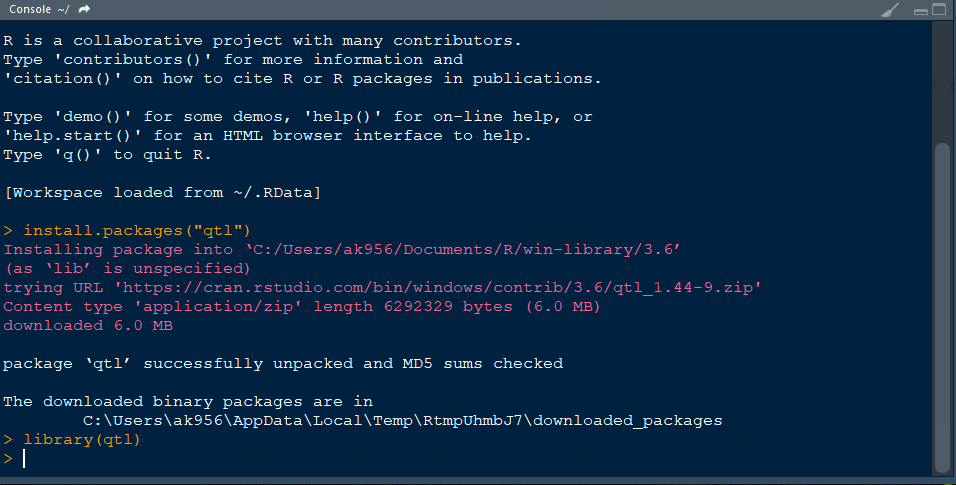
Install R package agricolae and open the library typing the below command line:
library(agricolae)
Note: Please remember to install the correct R package for ANOVA!
2.2 Import data
Import your data by typing the below command line:
data= read.table(file = "fileName.txt", header = T)
2.3 Check data
Once the data is imported, check it by typing the below command line:
head(data_pressure)
tail(data_pressure)
2.4 Conduct ANOVA
Now, Simply run ANOVA by typing the below command lines:
data.lm <- lm(data$Dependent_variable ~ data$Treatment, data = data)
data.av <- aov(data.lm)
summary(data.av)
2.5 Regression Coefficient
Obtain regression coefficient of the predictors in the data using below code:
summary(data.lm)
Output –>
> summary(data.lm)
Call:
lm(formula = data$Dependent_variable ~ data$Treatment, data = data)
Residuals:
Min 1Q Median 3Q Max
-2.32500 -0.48500 0.05917 0.23979 2.68500
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.9383 0.4481 4.325 0.000329 ***
data$TreatmentB 2.5267 0.6338 3.987 0.000726 ***
data$TreatmentD 5.7450 0.6338 9.065 1.61e-08 ***
data$TreatmentE 5.7258 0.6338 9.035 1.70e-08 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.098 on 20 degrees of freedom
Multiple R-squared: 0.8524, Adjusted R-squared: 0.8302
F-statistic: 38.49 on 3 and 20 DF, p-value: 1.692e-08
To add another coefficient, add the symbol “+” for every additional variable you want to add to the model.
2.6 Overall model’s performance
The overall model’s performane cand be obtained using the below code:
summary(data.av)
Output –>
> summary(data.av)
Df Sum Sq Mean Sq F value Pr(>F)
data$Treatment 3 139.2 46.38 38.49 1.69e-08 ***
Residuals 20 24.1 1.20
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
2.7 Good fit of the linear model
The coefficient of determination or R² is a good measure to test if the linear model has the good fit, and is measured by the proportion of the total variability explained by the regression model.
R² of a linear model can be obtained using below code:
summary(data.lm)$r.squared
Output –>
summary(data.lm)$r.squared
[1] 0.8523754
The model can explain ~85% of the total variability, which tells us that the model fits the data very well.
3.0 Conduct Tukey test
From the summary output, one can interpret that there is a significant difference (i.e. P < 0.001) between the Treatments, however, we perfom Tukey’s Test to investigate the differences between all treaments using steps below.
Type below commands to run Tukey test:
data.test <- TukeyHSD(data.av)
data.test
Below is the summary of the Tukey test:
> data.test
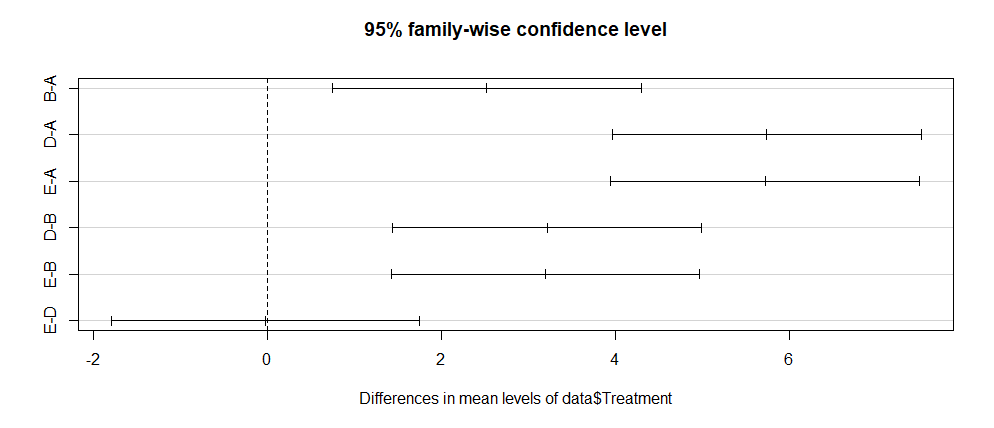
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = data.lm)
$`data$Treatment`
diff lwr upr p adj
B-A 2.52666667 0.7527896 4.300544 0.0037260
D-A 5.74500000 3.9711229 7.518877 0.0000001
E-A 5.72583333 3.9519563 7.499710 0.0000001
D-B 3.21833333 1.4444563 4.992210 0.0003106
E-B 3.19916667 1.4252896 4.973044 0.0003326
E-D -0.01916667 -1.7930437 1.754710 0.9999897
From the above T-test, one can conclude that there is a significant difference in the most of groups, except between-groups E-D at P <0.001
Finally, one can plot the above results using the below command:
plot(data.test)
Output:
--- End of Tutorial ---
Thank you for reading this tutorial. If you have any questions or comments, please let me know in the comment section below or send me an email.
Bibliography
Felipe de Mendiburu (2019). agricolae: Statistical Procedures for Agricultural Research. R package version 1.3-1. https://CRAN.R-project.org/package=agricolae